Dynamics of fluorescent proteins
In this project, we investigate various fluorescent proteins and other related
photoactive biomolecules. Studying these systems requires highly accurate descriptions
of the related potential energy surfaces, and this aspect has been the largest obstacle
in extending the timescales probed with any theoretical methods. Our group has developed
a new approach, named interpolated mechanics - molecular mechanics (IM/MM), for building
potential models with high accuracies and superb efficiencies. With this new scheme,
simulation time for a sizeable fluorescent protein can reach well over
a microsecond. We are continuously developing other approaches and are
implementing additional aspects that need to be included for correctly describing
excited state dynamics of protein systems. At the same time,
we are applying the approaches to studying detailed mechanisms
that control the behaviors of many photoproteins.
In fact, excited state dynamics in general has been puzzling many chemists for over decades already, and we still have much to learn to understand the fascinating phenomona in a satisfactory manner. Practically also, through such understanding, we can even have capabilities of suggesting directions for newly designing additional variations for many interesting photoprotein systems.
In fact, excited state dynamics in general has been puzzling many chemists for over decades already, and we still have much to learn to understand the fascinating phenomona in a satisfactory manner. Practically also, through such understanding, we can even have capabilities of suggesting directions for newly designing additional variations for many interesting photoprotein systems.
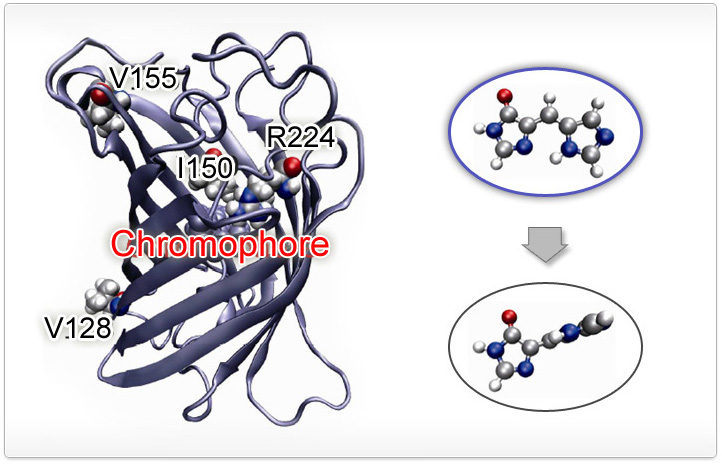 Chromophore dynamics in the electronically excited state leads to changes in emission
properties of many fluorescent proteins. For example, the blue fluorescent protein (BFP)
loses its emission when the chromophore twists as shown in the figure. Protein-chromophore
interaction can modulate this dynamics and accurate potential description can pin point
what factors contribute importantly for such modulations. For details, you can read
J. Phys. Chem. B 116, 11137 (2012).
Chromophore dynamics in the electronically excited state leads to changes in emission
properties of many fluorescent proteins. For example, the blue fluorescent protein (BFP)
loses its emission when the chromophore twists as shown in the figure. Protein-chromophore
interaction can modulate this dynamics and accurate potential description can pin point
what factors contribute importantly for such modulations. For details, you can read
J. Phys. Chem. B 116, 11137 (2012).
Quantum effects in photosynthetic systems
Until very recently, it was commonly believed that quantum behaviors such as
electronic coherence would have negligible effects in biological systems.
This is because any coherence would have an extremely short lifetime due to stochastic
fluctuations from the surrounding atoms.
However, long-lived electronic coherences among distant photosynthetic chromophores
have been repeatedly observed even at ambient temperatures. Because transfers of
excitons and electrons are important steps of many biologically
important processes, and because such steps will definitely involve
quantum mechanical aspects, we surely need to understand their governing principles.
In this project, we have been examining the detailed dynamics within photosynthetic complexes. Our major point of view is to include all atomistic details in the examination. For this, we have been treating a series of prototypical systems such as the famous Fenna-Matthews-Olson (FMO) complex. Treating the transfer dynamics with all-atom details actually requires different techniques than conventional classical mechanics based approaches (which are often called "MD" approaches). Thus, we apply nonadiabatic semiclassical techniques, and we are heavily developing practical and reliable schemes that can be newly applied for relevant systems. Ultimately, we would love to get to a stage where we can understand the dynamics of protein complexes or supercomplexes with many proteins, even in their embedding membrane. This will indeed be a huge challenge in various respects for quite some time in the future. (However, who doesn't love a challenge?!?) Through this study, we expect that we will be able to elucidate the fundamentals of quantum mechanical behaviors of various biological systems, and thus gain comprehensive pictures that we can use as a guidance in the future.
In this project, we have been examining the detailed dynamics within photosynthetic complexes. Our major point of view is to include all atomistic details in the examination. For this, we have been treating a series of prototypical systems such as the famous Fenna-Matthews-Olson (FMO) complex. Treating the transfer dynamics with all-atom details actually requires different techniques than conventional classical mechanics based approaches (which are often called "MD" approaches). Thus, we apply nonadiabatic semiclassical techniques, and we are heavily developing practical and reliable schemes that can be newly applied for relevant systems. Ultimately, we would love to get to a stage where we can understand the dynamics of protein complexes or supercomplexes with many proteins, even in their embedding membrane. This will indeed be a huge challenge in various respects for quite some time in the future. (However, who doesn't love a challenge?!?) Through this study, we expect that we will be able to elucidate the fundamentals of quantum mechanical behaviors of various biological systems, and thus gain comprehensive pictures that we can use as a guidance in the future.
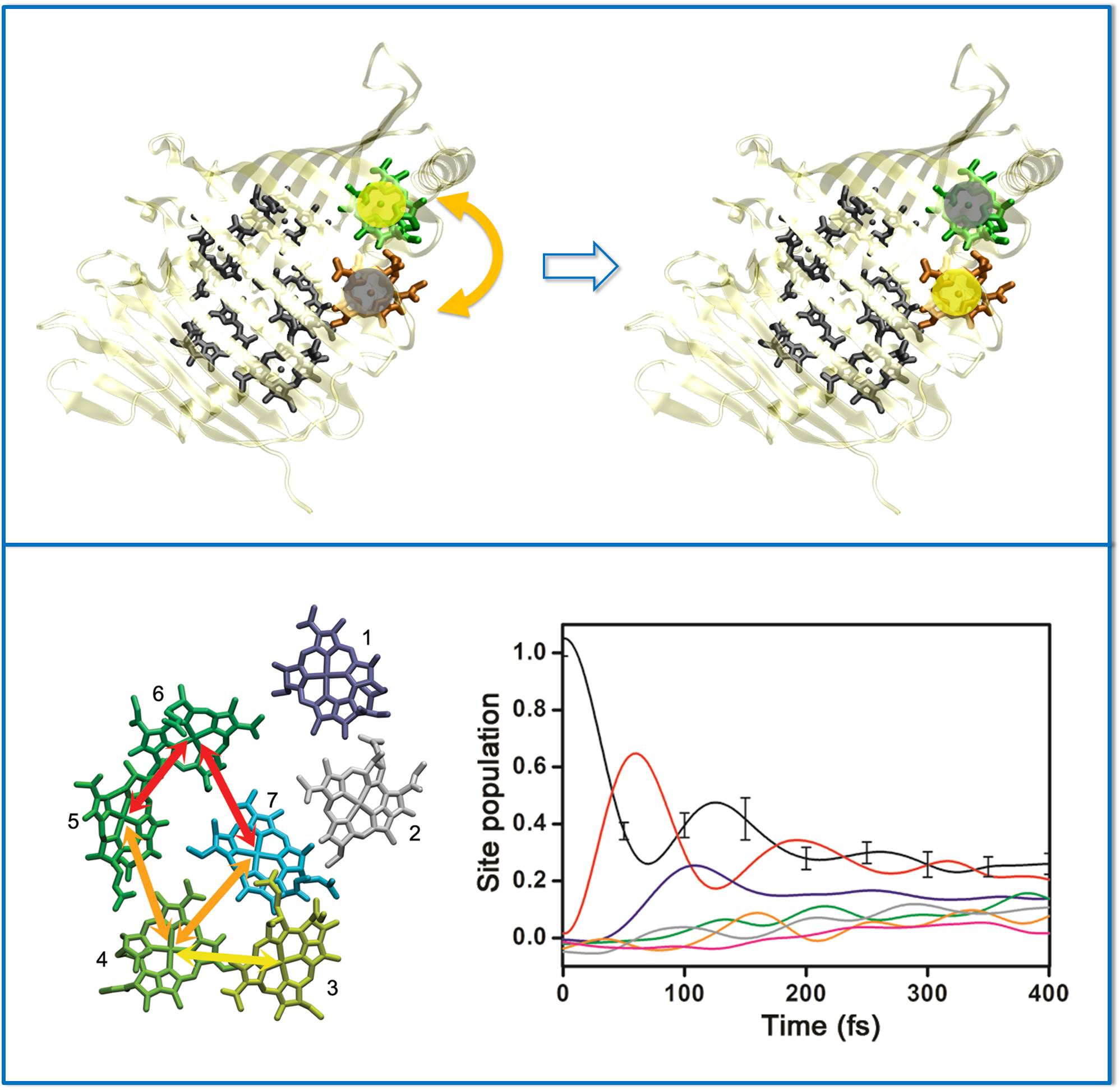 During the energy transfer process in the FMO complex (cartoon drawing at the top panel),
cross-talking between chromophores lasts for a long time even with noisy protein environment
(bottom panel).
Why and how that happens is a puzzle, and tackling that question with theoretical means
is also a challenge.
For details, you can read
J. Phys. Chem. Lett. 2, 808 (2011)
and
J. Am. Chem. Soc. 134, 11640 (2012).
During the energy transfer process in the FMO complex (cartoon drawing at the top panel),
cross-talking between chromophores lasts for a long time even with noisy protein environment
(bottom panel).
Why and how that happens is a puzzle, and tackling that question with theoretical means
is also a challenge.
For details, you can read
J. Phys. Chem. Lett. 2, 808 (2011)
and
J. Am. Chem. Soc. 134, 11640 (2012).
Bioluminescence: From a molecular view point
In this study, we aim to elucidate the detailed molecular picture of
the chromophore-protein interactions in bioluminescence processes.
Bioluminescence is found in many organisms and has recently shown important
promises as new in vivo cell imaging and therapeutic tools. While static structural
data of related luciferases are accumulating, their "movie-style" depictions
in their luminescing states are yet simply lacking.
If we can get such information, we will be able to attain better controls over the
application of bioluminescence. For example, luminescence color can be
modulated by protein modifications (without changing the luminophore at all!),
and protein-ligand interaction will definitely be the key to such
modifications. But people still do not know exactly with what mechanism
such modifications work.
In this project, we are attempting to analyze the chromophore-luciferase interactions and their effect on luminescence by combining both quantum chemical and statistical approaches. We also simulate the dynamics of the chromophore-luciferase complex to reveal the real picture of this interesting biochemical phenomenon. For this, we often develop models of interesting bioluminescent systems and perform molecular dynamics simulations. Interestingly, but not surprisingly, we almost always experience that chemical intuition is much more important than any model or any computation during the analysis of our data!
In this project, we are attempting to analyze the chromophore-luciferase interactions and their effect on luminescence by combining both quantum chemical and statistical approaches. We also simulate the dynamics of the chromophore-luciferase complex to reveal the real picture of this interesting biochemical phenomenon. For this, we often develop models of interesting bioluminescent systems and perform molecular dynamics simulations. Interestingly, but not surprisingly, we almost always experience that chemical intuition is much more important than any model or any computation during the analysis of our data!
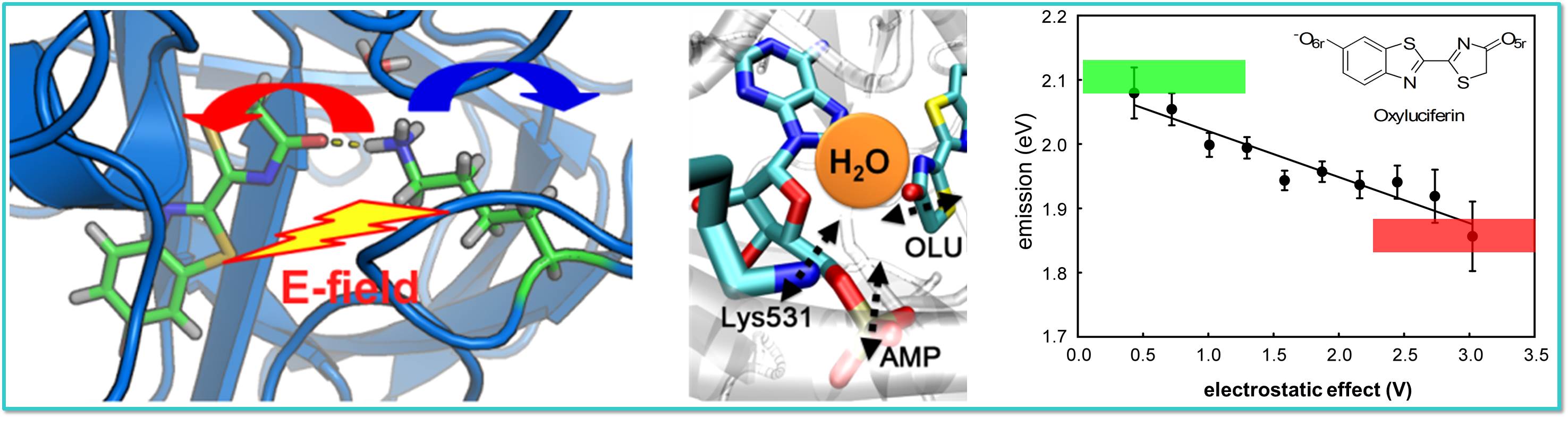 Oxyluciferin, the luminophore of firefly bioluminescence, can be affected by
various factors such as electrostatic interactions with neighboring protein
side chains and a water molecule in the active pocket of the luciferase enzyme.
One single type of oxyluciferin may emit from green to red with these factors.
Moreover, oxyluciferin may even be chemically modified
through tautomerization to attain a further diversified
repotoire of emission colors. Explaining its full detail is a challenging
but interesting topic in chemistry.
For details, you can read
J. Am. Chem. Soc. 133, 12040 (2011)
and
J. Phys. Chem. B 117, 7260 (2013)
.
Oxyluciferin, the luminophore of firefly bioluminescence, can be affected by
various factors such as electrostatic interactions with neighboring protein
side chains and a water molecule in the active pocket of the luciferase enzyme.
One single type of oxyluciferin may emit from green to red with these factors.
Moreover, oxyluciferin may even be chemically modified
through tautomerization to attain a further diversified
repotoire of emission colors. Explaining its full detail is a challenging
but interesting topic in chemistry.
For details, you can read
J. Am. Chem. Soc. 133, 12040 (2011)
and
J. Phys. Chem. B 117, 7260 (2013)
.